Authors: June Hidalgo, Unai Artetxe, José M. Becerril, María T. Gómez-Sagasti, Lur Epelde, Juan Vilela, Carlos Garbisu Journal: Environmental Science and Pollution Research DOI: 10.1007/s11356-023-31550-0 (more…)...
Read more...
Papers
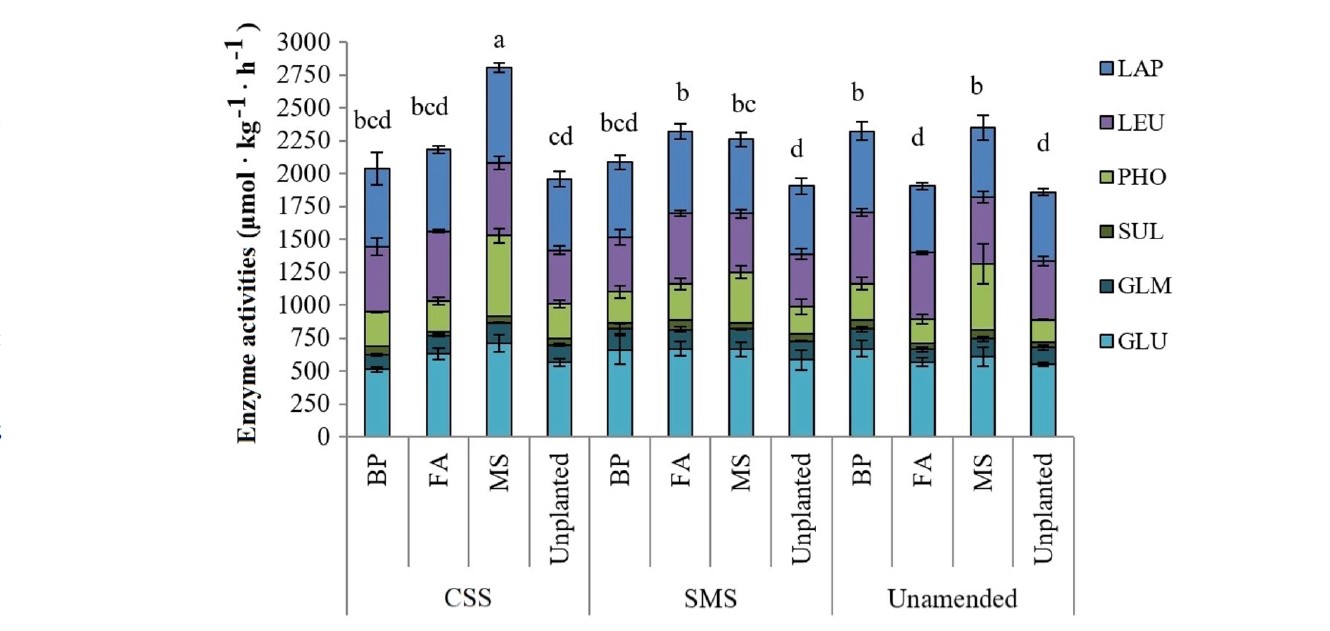
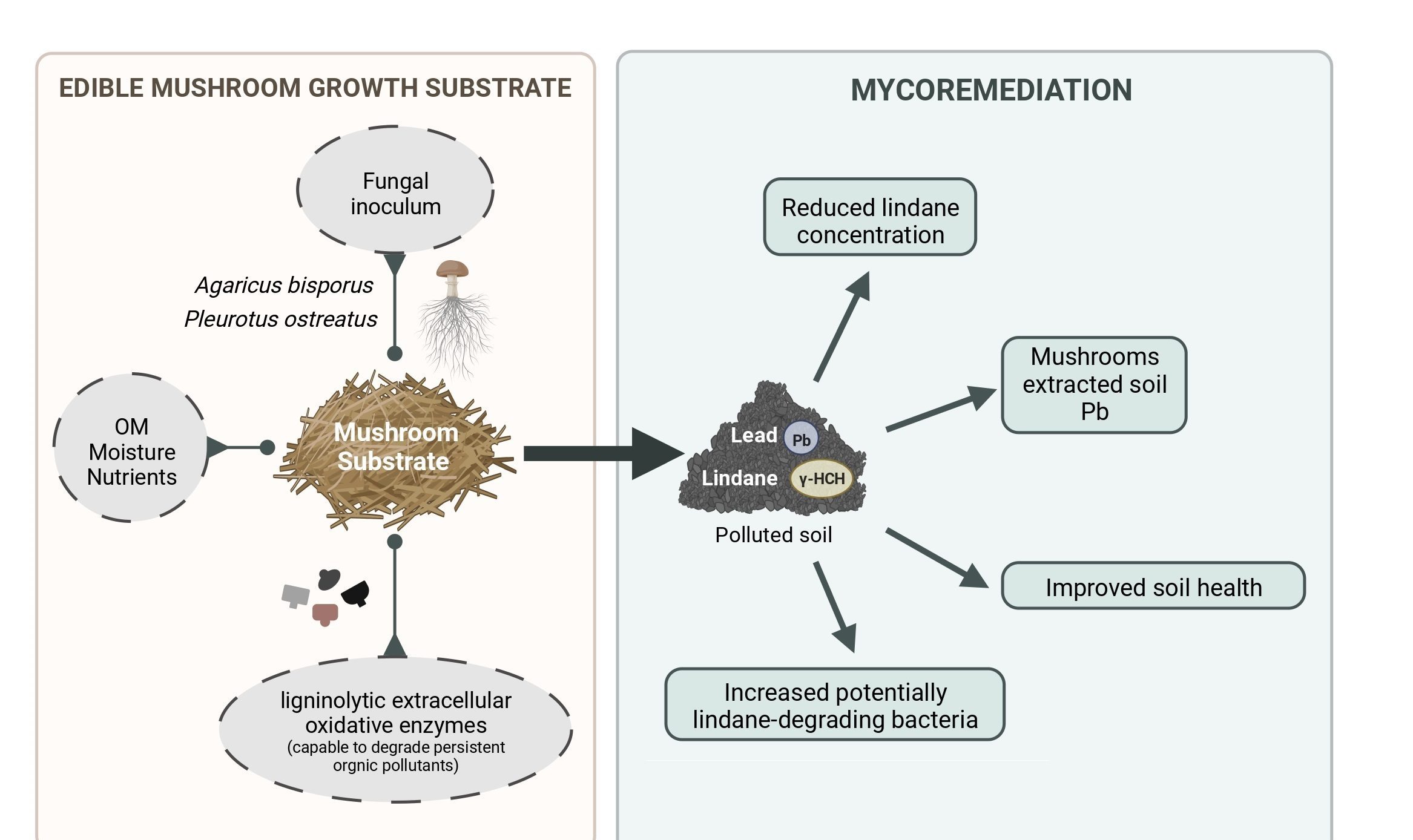
Mycoremediation with Agaricus bisporus and Pleurotus ostreatus growth substrates versus phytoremediation with Festuca rubra and Brassica sp. for the recovery of a Pb and γ-HCH contaminated soil
Authors: June Hidalgo, Lur Epelde, Mikel Anza, José M. Becerril, Carlos Garbisu Journal: Chemosphere https://doi.org/10.1016/j.chemosphere.2023.138538 (more…)...
Read more...
Read more...
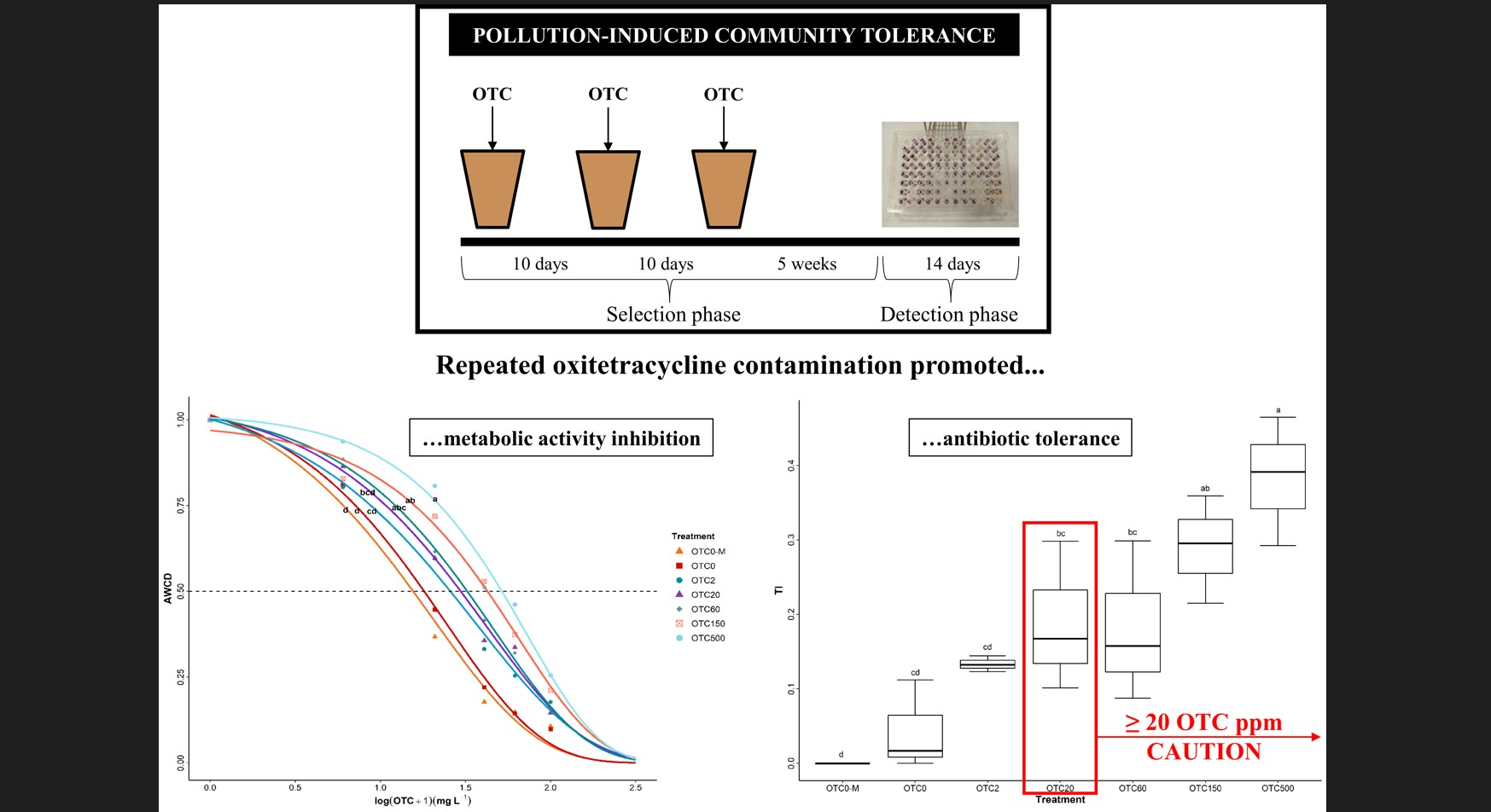
Induced development of oxytetracycline tolerance in bacterial communities from soil amended with well-aged cow manure
Authors: Leire Jauregi, Lur Epelde, Maddi Artamendi, Fernando Blanco, Carlos Garbisu Journal: Ecotoxicology https://rdcu.be/c9sNj (more…)...
Read more...
Read more...
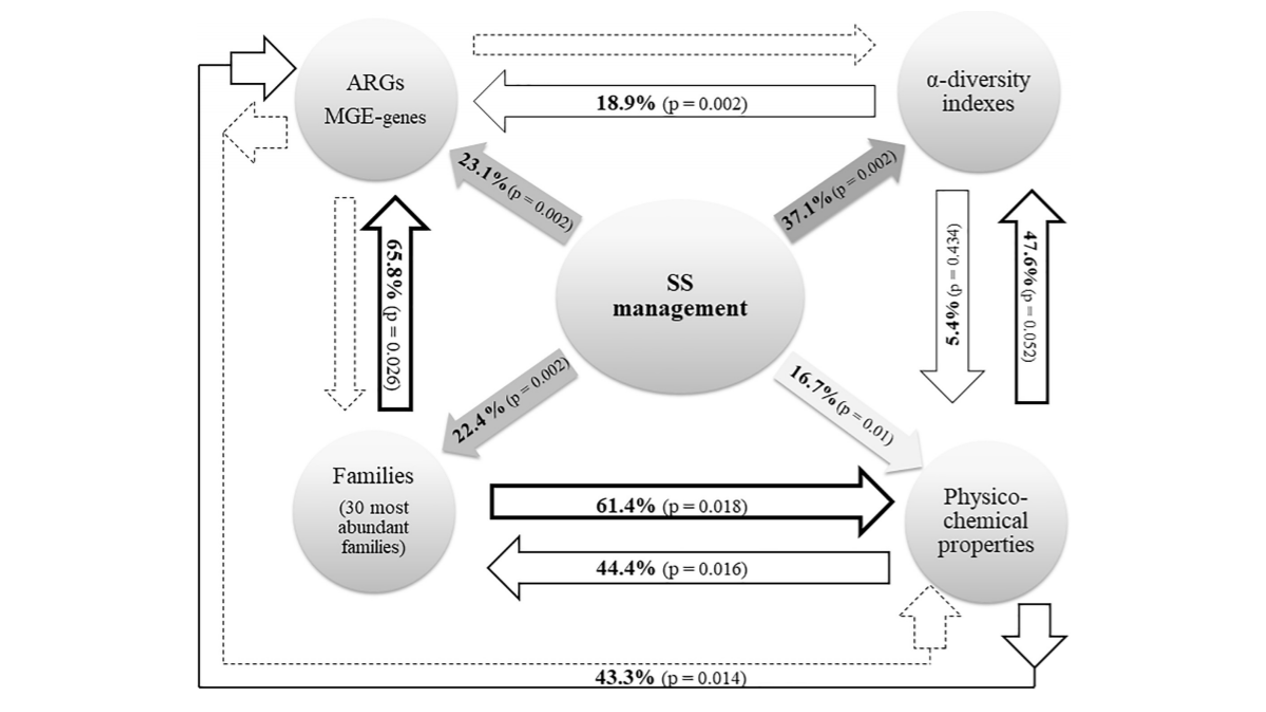
Organic amendment treatments for antimicrobial resistance and mobile element genes risk reduction in soil‑crop systems
Authors: Leire Jauregi, Aitor González , Carlos Garbisu, Lur Epelde Journal: Scientific Reports https://www.nature.com/articles/s41598-023-27840-9 (more…)...
Read more...
Read more...
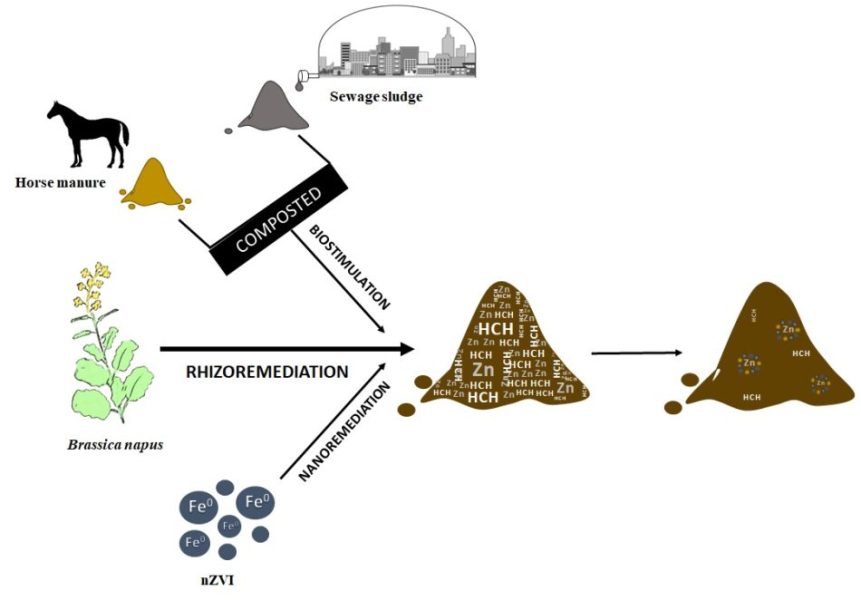
Zero-valent iron nanoparticles and organic amendment assisted rhizoremediation of mixed contaminated soil using Brassica napus
Authors: June Hidalgo, Mikel Anza, Lur Epelde, José M. Becerril, Carlos Garbisu Journal: Environmental Technology & Innovation https://doi.org/10.1016/j.eti.2022.102621 (more…)...
Read more...
Read more...
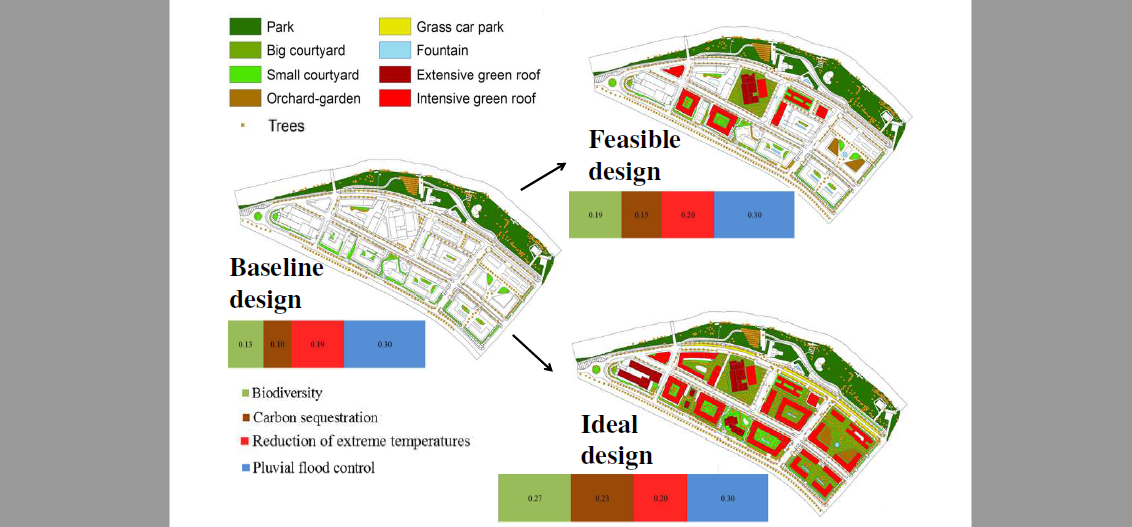
Quantification of the environmental effectiveness of nature-based solutions for increasing the resilience of cities under climate change
Authors: Lur Epelde, Maddalen Mendizabal, Laura Gutiérrez, Ainara Artetxe, Carlos Garbisu, Efrén Feliu Journal: Urban Forestry & Urban Greening https://doi.org/10.1016/j.ufug.2021.127433 (more…)...
Read more...
Read more...
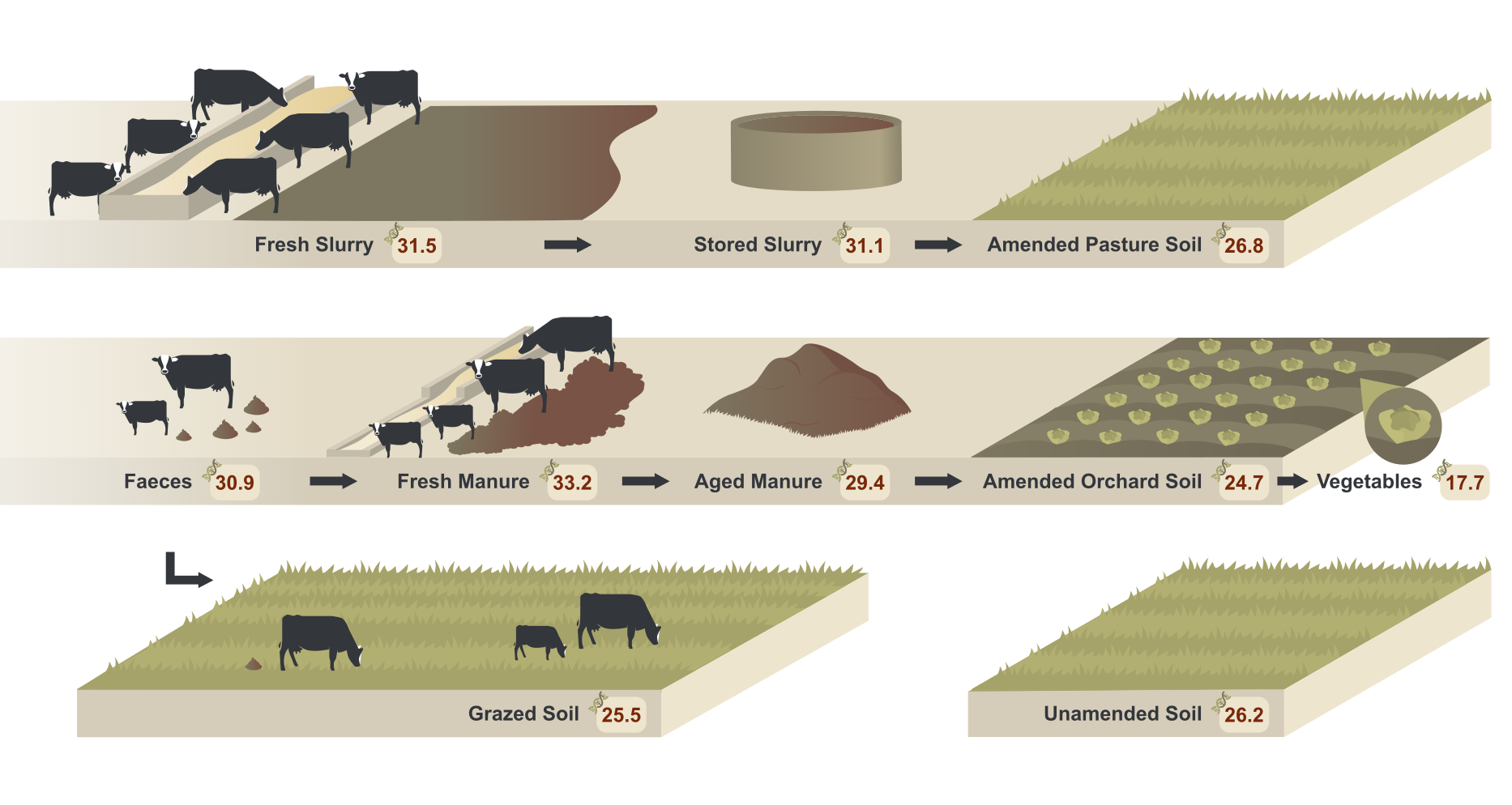
Reduction of the resistome risk from cow slurry and manure microbiomes to soil and vegetable microbiomes
Authors: Leire Jauregi, Lur Epelde, Aitor González, José Luis Lavín, Carlos Garbisu Journal: Environmental Microbiology https://doi.org/10.1111/1462-2920.15842 (more…)...
Read more...
Read more...
Agricultural soils amended with thermally-dried anaerobically-digested sewage sludge showed increased risk of antibiotic resistance dissemination
Authors: Leire Jauregi, Lur Epelde, Itziar Alkorta, Carlos Garbisu Journal: Frontiers in Microbiology Date: 2021 https://doi.org/10.3389/fmicb.2021.666854 (more…)...
Read more...
Read more...
Antibiotic resistance in agricultural soil and crops associated to the application of cow manure-derived amendments from conventional and organic livestock farms
Authors: Leire Jauregi, Lur Epelde, Itziar Alkorta, Carlos Garbisu Journal: Frontiers in Veterinary Science Date: 2021 https://doi.org/10.3389/fvets.2021.633858 (more…)...
Read more...
Read more...